Introduction
I will review the software tool article:
Crusoe MR, Alameldin HF, Awad S et al. The khmer software package: enabling efficient nucleotide sequence analysis [version 1; referees: 2 approved, 1 approved with reservations]. F1000Research 2015, 4:900 (doi: 10.12688/f1000research.6924.1)
I love this paper for several interrelated reasons:
- I am an author
- The paper is controversial
- Part of the controversy is whether or not I should be an author
Birney E. Referee Report For: The khmer software package: enabling efficient nucleotide sequence analysis [version 1; referees: 2 approved, 1 approved with reservations]. F1000Research 2015, 4:900 (doi: 10.5256/f1000research.7456.r10514)
Methods
The methods section of the paper describes how the software is implemented and used.
The software has a core C++ library for k-mer counting and graph traversal. The core is embeded into python with some custom C code. Most of the user facing software is composed of python scripts that call into the core libraries. khmer makes use of SeqAn's C++ parsers for sequence files.
Most of the algorithms used by khmer are described in other papers. I'll show some methods below.
The use of khmer is described in detail online http://khmer.readthedocs.io/en/v2.0/
Use cases
Digital Normalization
This was how I first learned about khmer. Titus has given several workshops, including ours, where he runs digital normalization on RNASeq data before assembly.
In fact, one of my contributions to khmer is a one line patch to the digital normalization script.
https://github.com/dib-lab/khmer/blob/v2.0/scripts/normalize-by-median.py#L162
The method of digital normalization is described in this preprint:
Brown CT, Howe A, Zhang Q, et al.: A reference-free algorithm for computational normalization of shotgun sequencing data. arXiv preprint. 2012. Reference Source http://arxiv.org/pdf/1203.4802v2.pdf
Which cites this paper for the underlying data structure:
Cormode G, Muthukrishnan S (2004) An improved data stream summary: the count-min sketch, and its applications. Journal of Algorithms http://dimacs.rutgers.edu/~graham/pubs/papers/cm-full.pdf
Figure 1 from the above paper shows how count-min sketch works.
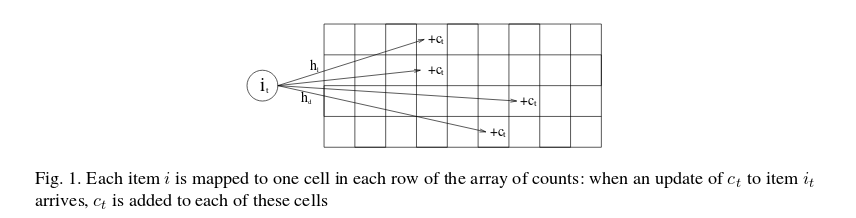
The Count Min sketch makes use of pairwise-independent families of functions for hashing, which I had to google:
Broder, Andrei Z., et al. "Min-wise independent permutations." Proceedings of the thirtieth annual ACM symposium on Theory of computing. ACM, 1998. https://www.princeton.edu/~rblee/ELE572Papers/Fall04Readings/Misc/minwise.pdf
K-mer counting and read trimming
See:
Zhang Q, Pell J, Canino-Koning R, et al.: These are not the k-mers you are looking for: Efficient online k-mer counting using a probabilistic data structure. PLoS One. 2014; 9(7): e101271. http://dx.doi.org/10.1371/journal.pone.0101271
Graph partitioning
Howe AC, Jansson JK, Malfatti SA, et al.: Tackling soil diversity with the assembly of large, complex metagenomes. Proc Natl Acad Sci U S A. 2014; 111(13): 4904–9. http://www.ncbi.nlm.nih.gov/pmc/articles/3977251
Discussion
The paper presents several practical applications of k-mer analysis of next-generation sequencing data. These applications are implemented in a mature software package, the khmer-tools. It serves as a good starting point for learning about k-mers and de Brujin graphs.
As is typical of journal papers, to really understand it you need to read many other papers, by digging through the references.
The khmer-tools development community is very open, and accepted my contributions to the code quickly, even including me as an author on this paper due to my small contribution to the code.