Review: Locality sensitive hashing for the edit distance
I'm reviewing some of the findings from the preprint:
Locality sensitive hashing for the edit distance Guillaume Marcais, Dan DeBlasio, Prashant Pandey, Carl Kingsford bioRxiv 534446; doi: https://doi.org/10.1101/534446
Problem
Determine if two sequences overlap.
Existing solutions
Locality sensitive hashing:
- LSH Jaccard similarity
- LSH Hamming distance
Proposal
Develop a LSH for edit distance.
Hamming distance
S1 GTACACTA
S2 TTATACTG
--------
x x x
Hd(S1, S2) = 3 / 8
Similarity
S = 1 - D
Jaccard similarity
S1 {GTA TAC ACA CAC ACT CTA}
S2 {TTA TAT ATA TAC ACT CTG}
{S1 n S2} = {ACT}
{S1 U S2} = {GTA TAC ACA CAC ACT CTA TTA TAT ATA TAC CTG}}
J(S1, S2) = |S1 n S2|/|S1 U S2| = 1/11
Edit distance
S1 GTACACTA
S2 TTAACTA
Ed(S1,S2) = 2/8
Locality Sensitive Hash
Gapped LSH
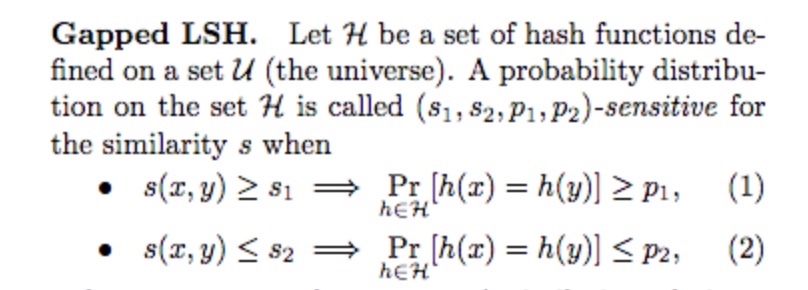
LSH

Order MinHash
A concrete example of Figure 1, with m = GTA and k = 3.
GTACACGTATGTAC {(GTA 0) (GTA 1) (GTA 2)}
--- --- ---
TCGTATGTAC {(GTA 0) (GTA 1)}
--- ---